Metagenomic analysis to identify gene clusters associated with IgA production
Project Information
bioinformatics, biology, genomics, python, r, research-facilitationProject Status: Finishing Up
Project Region: CAREERS
Submitted By: Joel Wilmore
Project Email: wilmorej@upstate.edu
Project Institution: SUNY Upstate Medical University
Anchor Institution: CR-Rensselaer Polytechnic Institute
Project Address: Syracuse , New York
Mentors: Joel Wilmore, Neil McGlohon
Students: Eren Ada
Project Description
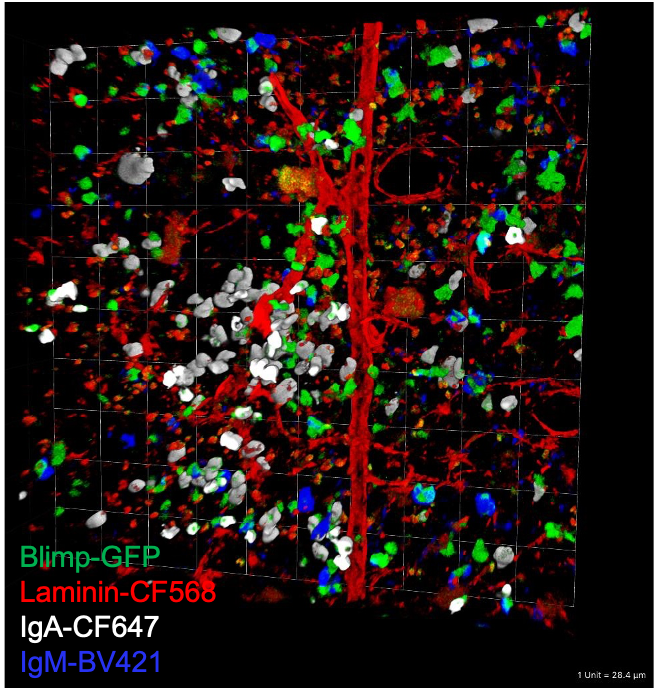
The development of systemic IgA responses is highly dependent on the composition of the intestinal microbiome. We performed shotgun metagenomics on fecal bacteria based on the binding of serum IgA (IgA+ vs IgA-). Our goal is to identify genes or functional clusters of genes that are highly associated with serum IgA binding. We will then be able to target specific pathways, rather than specific species or classes of microbes. Identification of these pathways will provide the basis for improved mucosal vaccine design or inflammatory bowel disease treatments.
Project Information
bioinformatics, biology, genomics, python, r, research-facilitationProject Status: Finishing Up
Project Region: CAREERS
Submitted By: Joel Wilmore
Project Email: wilmorej@upstate.edu
Project Institution: SUNY Upstate Medical University
Anchor Institution: CR-Rensselaer Polytechnic Institute
Project Address: Syracuse , New York
Mentors: Joel Wilmore, Neil McGlohon
Students: Eren Ada