Understanding Mechanical Properties of Bio-interfaces with HPC Molecular Simulations
Project Information
bioinformatics, computational-chemistry, gromacs, molecular-dynamics, mpi, pythonProject Status: Complete
Project Region: Northeast
Submitted By: Juan Vanegas
Project Email: jvanegas@uvm.edu
Project Institution: University of Vermont
Anchor Institution: NE-University of Vermont
Project Address: 82 University Place
Discovery Bldg W428
Burlington, Vermont. 05408
Students: Ali Razavi
Project Description
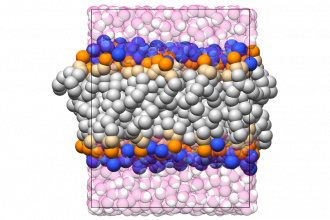
Molecular simulations are emerging as a new form of “microscopy” that can uniquely probe the behavior of biomolecules and interfacial systems such as biomembranes at the nanometer scale. High resolution techniques such as classical and first principles molecular dynamics (MD) can be used to understand the connection between molecular structure, biological function, and mechanical properties of biomaterials. In this project, students will use high performance MD simulation engines such as GROMACS (www.gromacs.org) to model biological interfaces and use custom local stress codes to characterize the mechanical properties of these systems.
Project Information
bioinformatics, computational-chemistry, gromacs, molecular-dynamics, mpi, pythonProject Status: Complete
Project Region: Northeast
Submitted By: Juan Vanegas
Project Email: jvanegas@uvm.edu
Project Institution: University of Vermont
Anchor Institution: NE-University of Vermont
Project Address: 82 University Place
Discovery Bldg W428
Burlington, Vermont. 05408
Students: Ali Razavi